Journal club
We host a journal club once every two weeks on Wednesdays at 11.30-12.30 am. A member of the Landry lab will select a single paper of interest and then discuss it with the attendees. The journal club is very informal, and all members of IBIS are welcome to attend!
If you would like to subscribe to the mailing list for this journal club, please follow the instructions here.
Please direct any queries regarding the journal club to: Simon Aubé
Wednesday 26th September, 2022
Mutation bias reflects natural selection in Arabidopsis thaliana
J Grey Monroe, Thanvi Srikant, Pablo Carbonell-Bejerano, Claude Becker, Mariele Lensink, Moises Exposito-Alonso, Marie Klein, Julia Hildebrandt, Manuela Neumann, Daniel Kliebenstein, Mao-Lun Weng, Eric Imbert, Jon Ågren, Matthew T Rutter, Charles B Fenster, Detlef Weigel
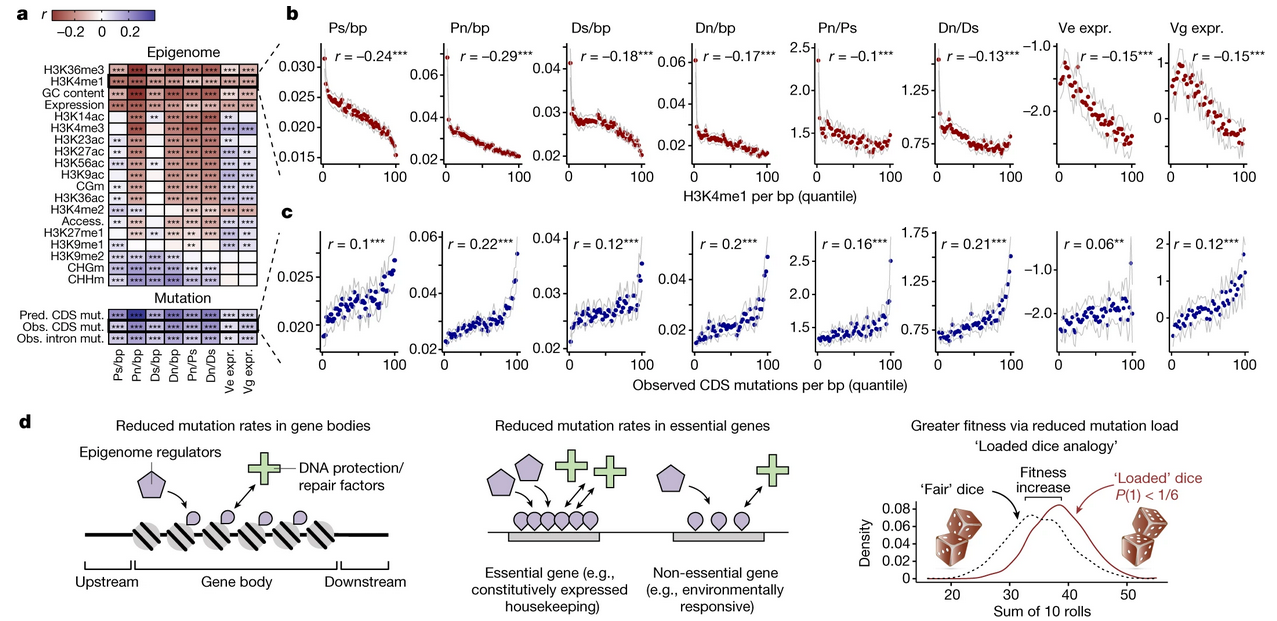
Nature
Responsible for the discussion: Simon Aubé
Tuesday 19th April, 2022
Contingency and selection in mitochondrial genome dynamics
Christopher J Nunn, Sidhartha Goyal
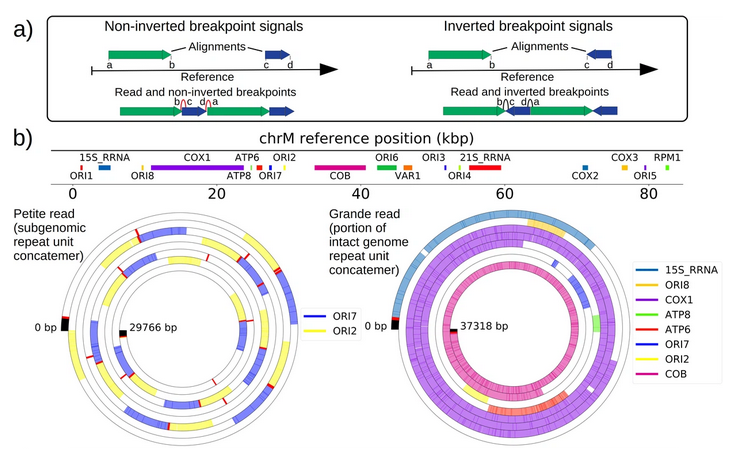
eLife
Responsible for the discussion: Damien Biot-Pelletier
Tuesday 5th April, 2022
Expression level is a major modifier of the fitness landscape of a protein coding gene
Zhuoxing Wu, Xiujuan Cai, Xin Zhang, Yao Liu, Guo-bao Tian, Jian-Rong Yang, Xiaoshu Chen
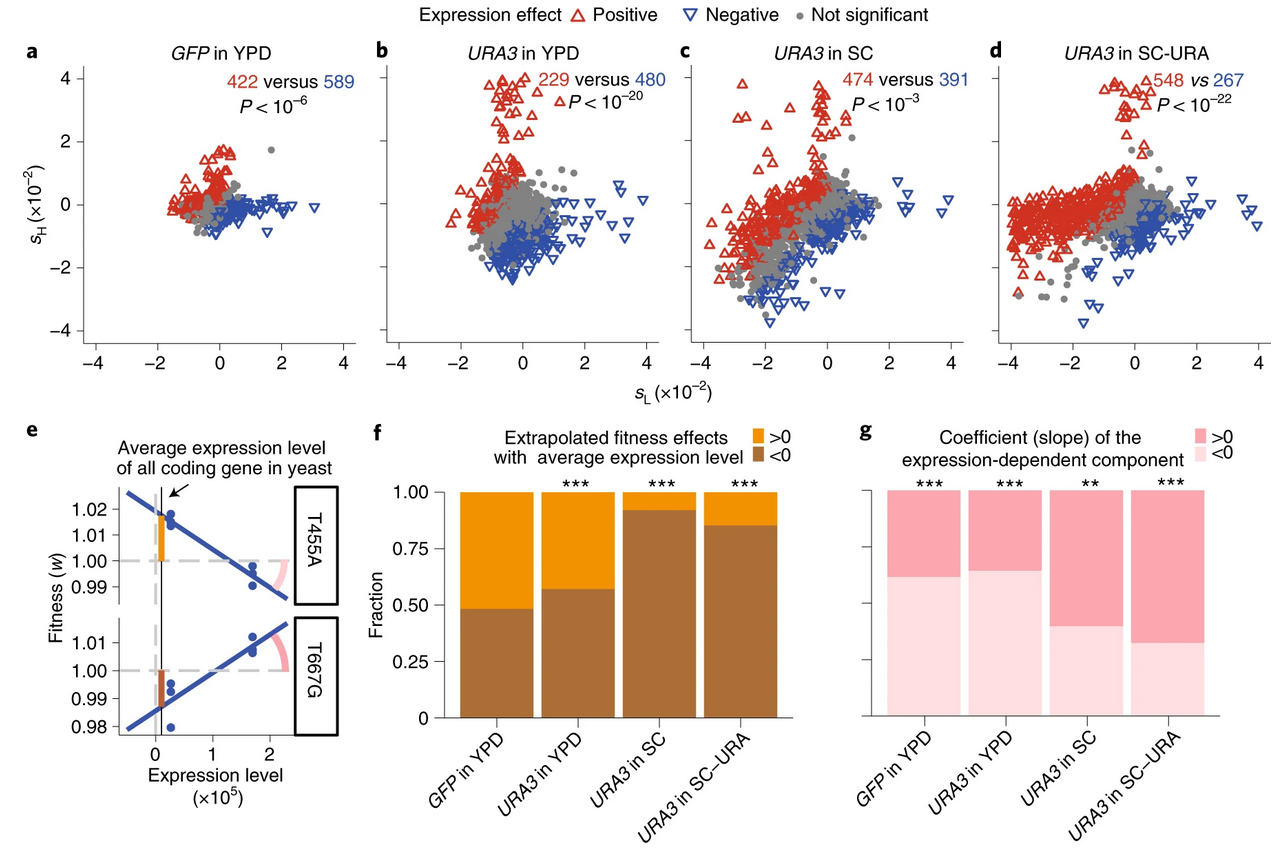
Nature Ecology & Evolution
Responsible for the discussion: Angel Cisneros
Tuesday 22nd March, 2022
Domestication of the Emblematic White Cheese-Making Fungus Penicillium camemberti and Its Diversification into Two Varieties
Jeanne Ropars, Estelle Didiot, Ricardo C. Rodríguez de la Vega, Bastien Bennetot, Monika Coton, Elisabeth Poirier, Emmanuel Coton, Alodie Snirc, Stéphanie Le Prieur, Tatiana Giraud
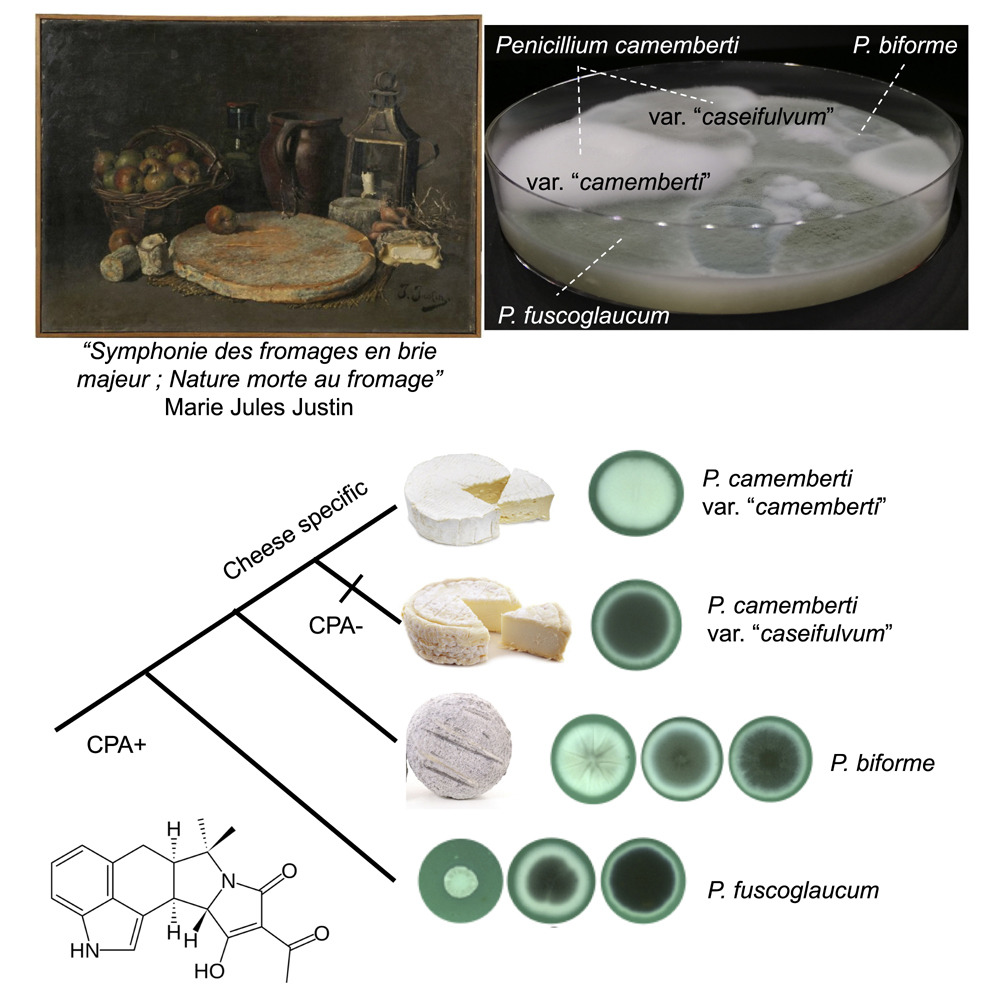
Current Biology
Responsible for the discussion: Emilie Alexander
Wednesday 9th March, 2022
Acquisition of cross-azole tolerance and aneuploidy in Candida albicans strains evolved to posaconazole
Rebekah J. Kukurudz, Madison Chapel, Quinn Wonitowy, Abdul-Rahman Adamu Bukari, Brooke Sidney, Riley Sierhuis, Aleeza C. Gerstein
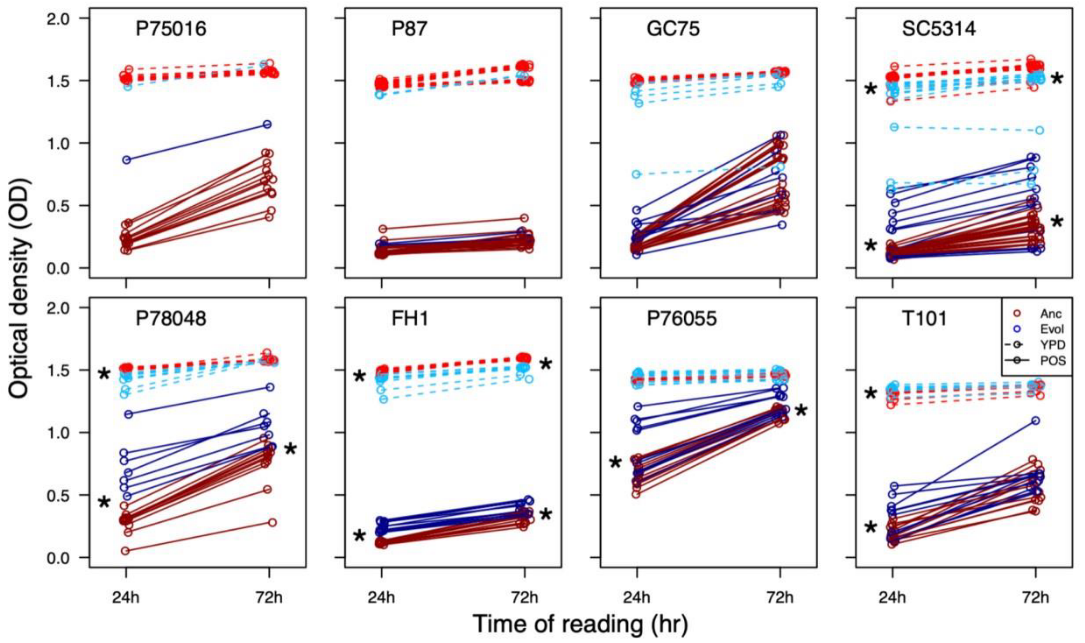
bioRxiv
Responsible for the discussion: Camille Bédard
Tuesday 22nd February, 2022
Co-option of the same ancestral gene family gave rise to mammalian and reptilian toxins
Agneesh Barrua, Ivan Koludarov, Alexander S. Mikheyev
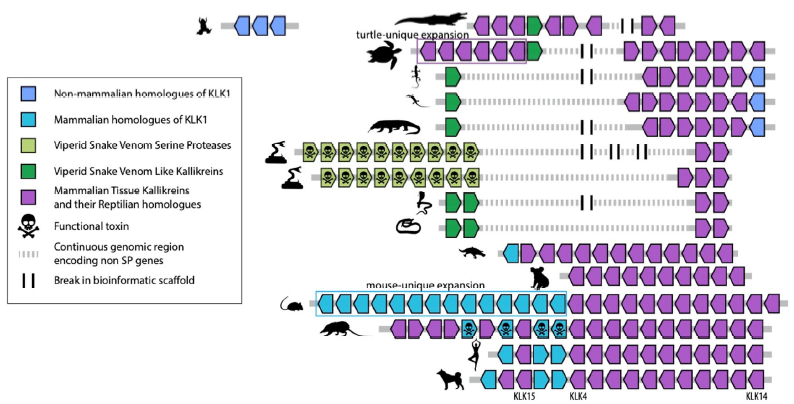
BMC Biology
Responsible for the discussion: Pascale Lemieux
Tuesday 8th February, 2022
A computational exploration of resilience and evolvability of protein–protein interaction networks
Brennan Klein, Ludvig Holmér, Keith M Smith, Mackenzie M Johnson, Anshuman Swain, Laura Stolp, Ashley I Teufel, April S Kleppe
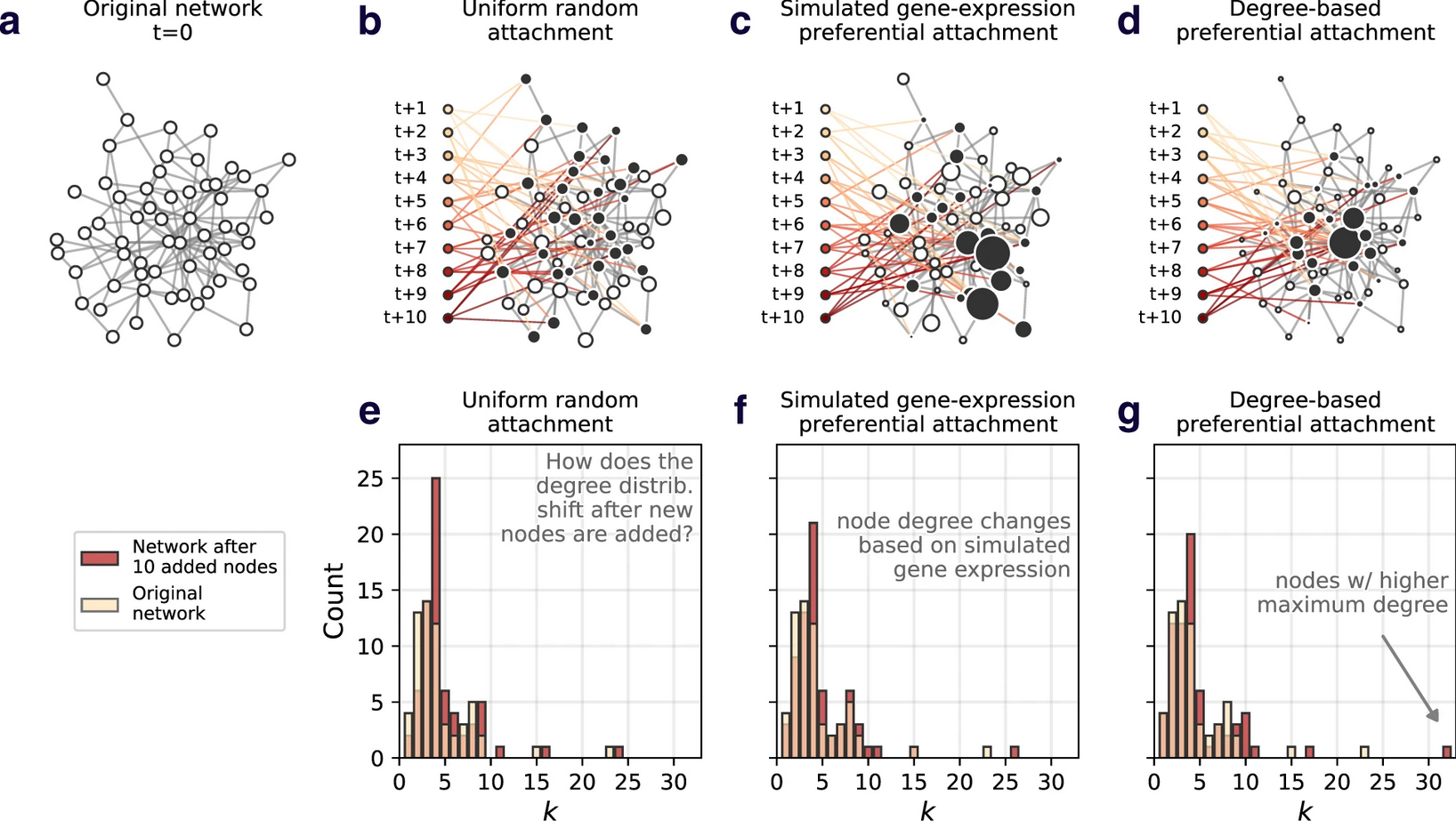
Communications Biology
Responsible for the discussion: Simon Aubé
Tuesday 25th January, 2022
Climbing Up and Down Binding Landscapes through Deep Mutational Scanning of Three Homologous Protein-Protein Complexes
Michael Heyne, Jason Shirian, Itay Cohen, Yoav Peleg, Evette S. Radisky, Niv Papo, Julia M. Shifman
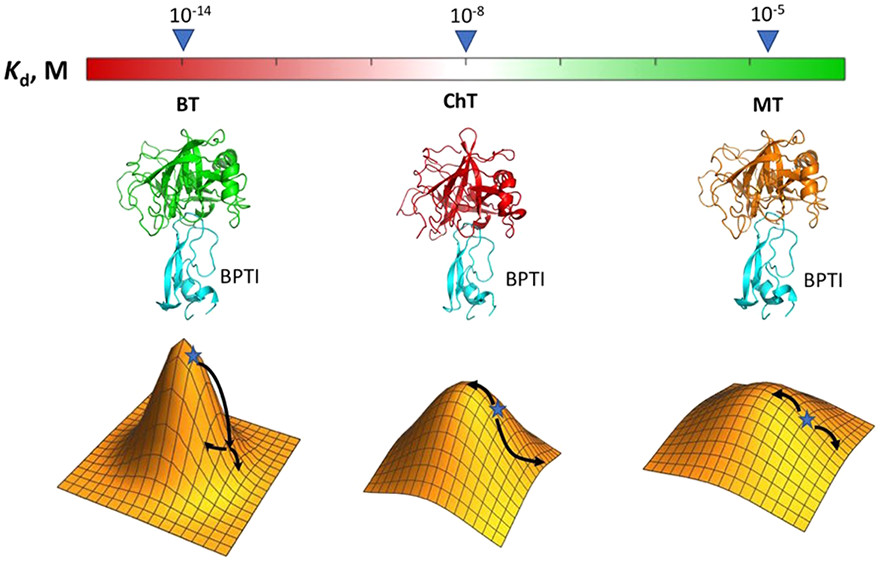
J. Am. Chem. Soc.
Responsible for the discussion: Soham Dibyachintan
Tuesday 11th January, 2022
An in vitro system to silence mitochondrial gene expression
Luis Daniel Cruz -Zaragoza, Sven Dennerlein, Andreas Linden, Roya Yousefi, Elena Lavdovskaia, Abhishek Aich, Rebecca R.Falk, Ridhima Gomkale, Thomas Schöndorf, Markus T.Bohnsack, Ricarda Richter-Dennerlein, Henning Urlaub, Peter Rehling
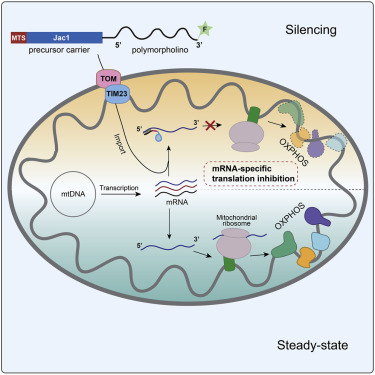
doi: https://doi.org/10.1016/j.cell.2021.09.033
Cell
Responsible for the discussion: Damien Biot-Pelletier
Wednesday 8th December, 2021
Herpesviruses assimilate kinesin to produce motorized viral particles
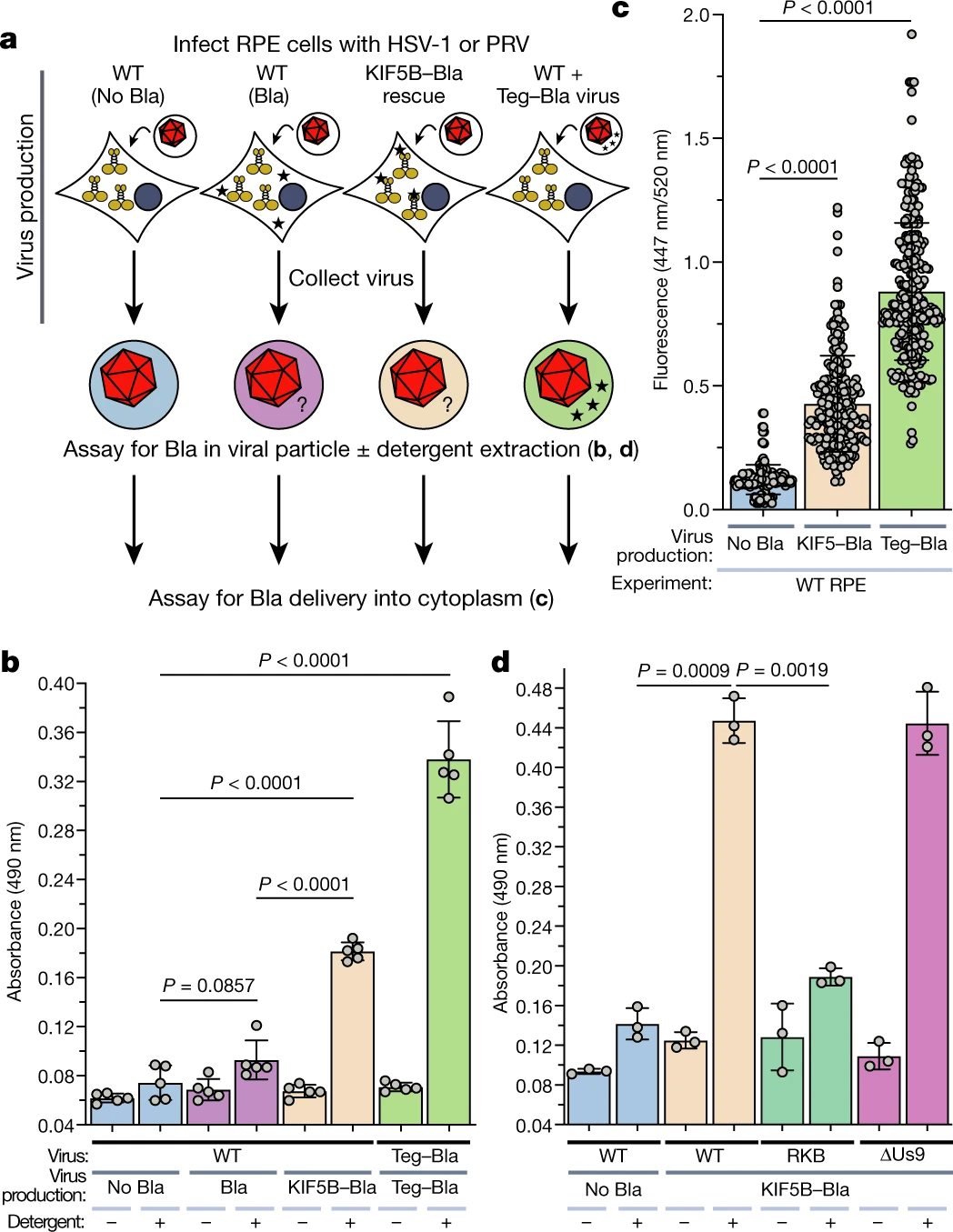
doi: https://doi.org/10.1038/s41586-021-04106-w
Nature
Responsible for the discussion: David Jordan
Wednesday 24th November, 2021
The structure of genotype-phenotype maps makes fitness landscapes navigable
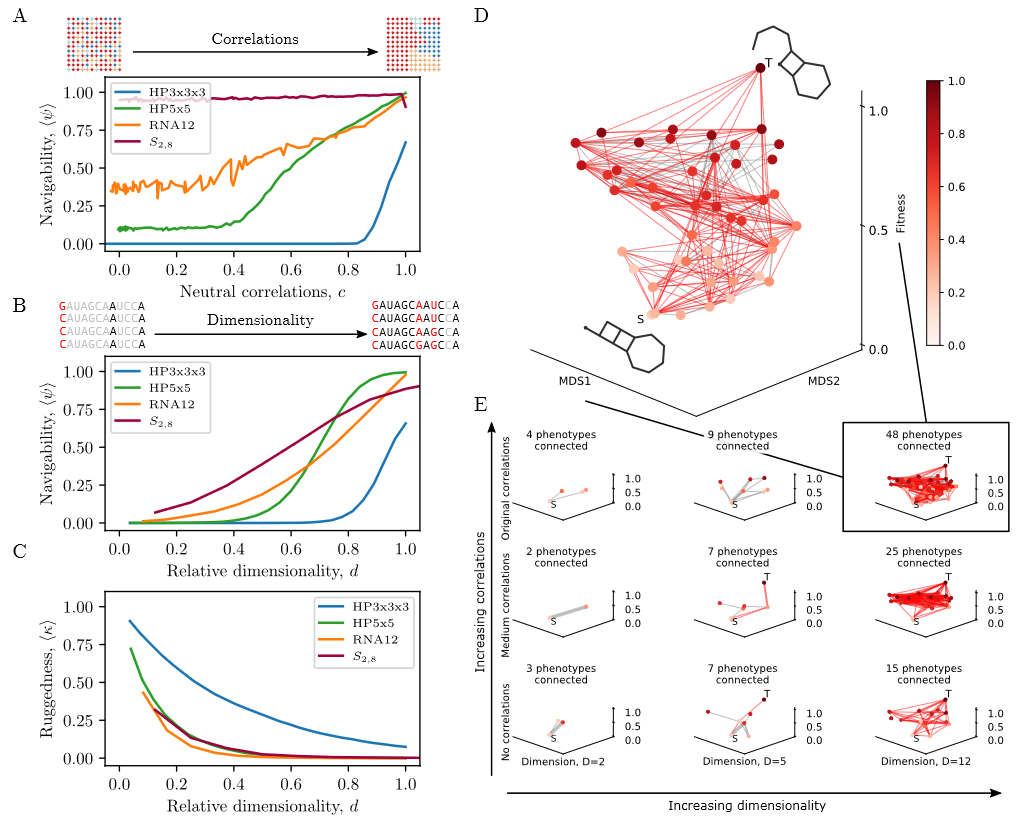
doi: https://doi.org/10.1101/2021.10.11.463990
bioRxiv
Responsible for the discussion: Alicia Pageau
Wednesday 10th November, 2021
Circular RNA repertoires are associated with evolutionarily young transposable elements
Franziska Gruhl, Peggy Janich, Henrik Kaessmann, David Gatfield
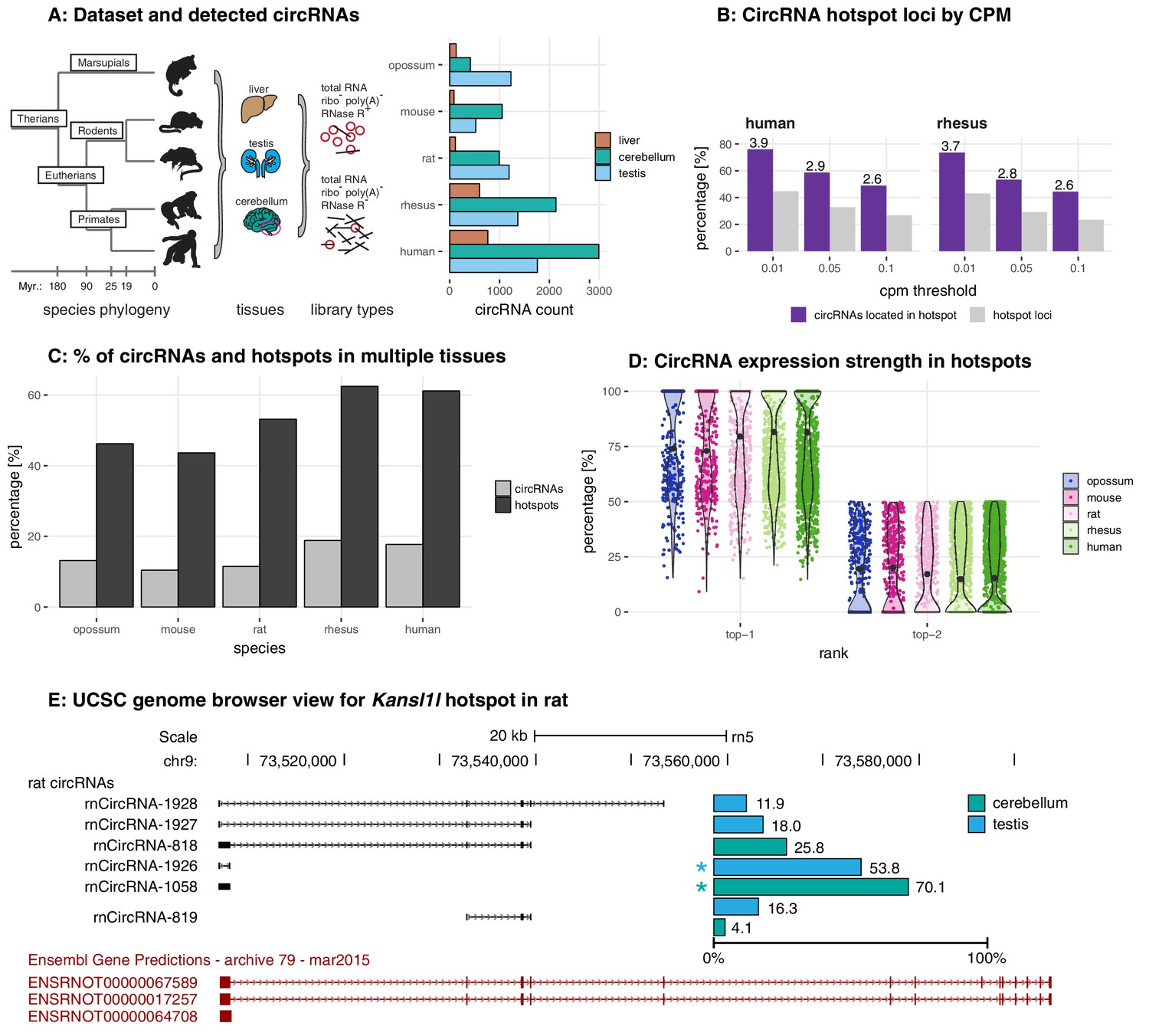
doi: https://elifesciences.org/articles/67991
eLife
Responsible for the discussion: Daniel Evans-Yamamoto
The lab
The Landry lab is located at the Institut de Biologie Intégrative et des Systèmes (IBIS) of Université Laval and is part of the Quebec Network for Research on Protein Function, Engineering, and Applications (PROTEO). IBIS and PROTEO offer very stimulating training environments and cutting edge technologies. The Landry lab is an international team of about 30 students, PDFs and research associates from different backgrounds (microbiology, biology, bioinformatics, biochemistry) addressing questions in evolutionary cell and systems biology.
The lab